




GluR-2 rabbit pAb
 One-click to copy product information
One-click to copy product information$148.00/50µL $248.00/100µL
| 50 µL | $148.00 |
| 100 µL | $248.00 |
Overview
| Product name: | GluR-2 rabbit pAb |
| Reactivity: | Human;Mouse;Rat |
| Alternative Names: | GRIA2; GLUR2; Glutamate receptor 2; GluR-2; AMPA-selective glutamate receptor 2; GluR-B; GluR-K2; Glutamate receptor ionotropic; AMPA 2; GluA2 |
| Source: | Rabbit |
| Dilutions: | Western Blot: 1/500 - 1/2000. Immunohistochemistry: 1/100 - 1/300. Immunofluorescence: 1/200 - 1/1000. ELISA: 1/20000. Not yet tested in other applications. |
| Immunogen: | The antiserum was produced against synthesized peptide derived from human GluR2. AA range:834-883 |
| Storage: | -20°C/1 year |
| Clonality: | Polyclonal |
| Isotype: | IgG |
| Concentration: | 1 mg/ml |
| Observed Band: | 99kD |
| GeneID: | 2891 |
| Human Swiss-Prot No: | P42262 |
| Cellular localization: | Cell membrane ; Multi-pass membrane protein . Endoplasmic reticulum membrane ; Multi-pass membrane protein . Cell junction, synapse, postsynaptic cell membrane ; Multi-pass membrane protein . Cell junction, synapse, postsynaptic density membrane ; Multi-pass membrane protein . Interaction with CACNG2, CNIH2 and CNIH3 promotes cell surface expression (By similarity). Displays a somatodendritic localization and is excluded from axons in neurons (By similarity). . |
| Background: | Glutamate receptors are the predominant excitatory neurotransmitter receptors in the mammalian brain and are activated in a variety of normal neurophysiologic processes. This gene product belongs to a family of glutamate receptors that are sensitive to alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA), and function as ligand-activated cation channels. These channels are assembled from 4 related subunits, GRIA1-4. The subunit encoded by this gene (GRIA2) is subject to RNA editing (CAG->CGG; Q->R) within the second transmembrane domain, which is thought to render the channel impermeable to Ca(2+). Human and animal studies suggest that pre-mRNA editing is essential for brain function, and defective GRIA2 RNA editing at the Q/R site may be relevant to amyotrophic lateral sclerosis (ALS) etiology. Alternative splicing, resulting in transcript variants enco |
-
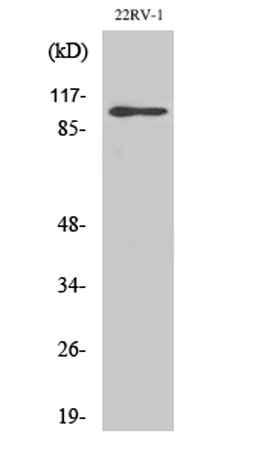
 Western Blot analysis of various cells using GluR-2 Polyclonal Antibody
Western Blot analysis of various cells using GluR-2 Polyclonal Antibody -
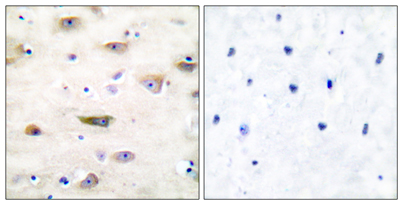
 Immunohistochemistry analysis of paraffin-embedded human brain tissue, using GluR2 Antibody. The picture on the right is blocked with the synthesized peptide.
Immunohistochemistry analysis of paraffin-embedded human brain tissue, using GluR2 Antibody. The picture on the right is blocked with the synthesized peptide. -
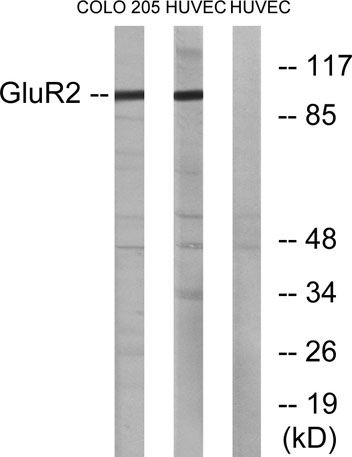
 Western blot analysis of lysates from COLO205 and HUVEC cells, using GluR2 Antibody. The lane on the right is blocked with the synthesized peptide.
Western blot analysis of lysates from COLO205 and HUVEC cells, using GluR2 Antibody. The lane on the right is blocked with the synthesized peptide.

 Manual
Manual